项目:fasta 转化 md5库
工作中的一个小的细节记录
将 fasta 用 md5 统一 id
- 拿到 md5 串
- 去除第1行
>开头的信息 - 将后面行,连接
- 去掉
\n,去2侧的空格 - 转成纯大写
fasta 串
这个串在 .fasta 文件里是严格行数显示的,所以不用担心一些奇奇怪怪的问题。
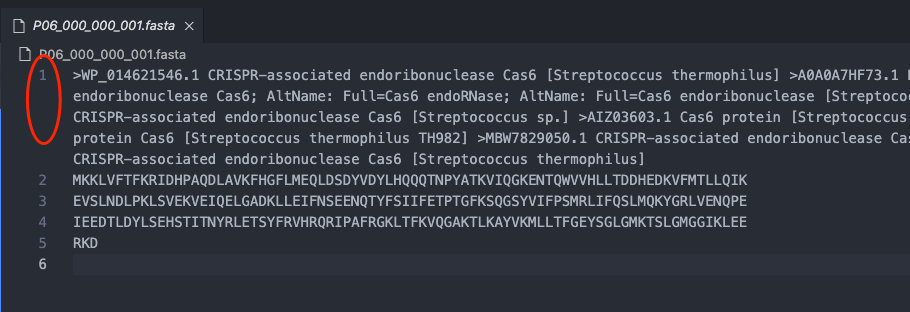
>WP_014621546.1 CRISPR-associated endoribonuclease Cas6 [Streptococcus thermophilus] >A0A0A7HF73.1 RecName: Full=CRISPR-associated endoribonuclease Cas6; AltName: Full=Cas6 endoRNase; AltName: Full=Cas6 endoribonuclease [Streptococcus thermophilus] >MBR2538257.1 CRISPR-associated endoribonuclease Cas6 [Streptococcus sp.] >AIZ03603.1 Cas6 protein [Streptococcus thermophilus] >EWM58862.1 CRISPR-associated protein Cas6 [Streptococcus thermophilus TH982] >MBW7829050.1 CRISPR-associated endoribonuclease Cas6 [Streptococcus thermophilus] >MCA6641324.1 CRISPR-associated endoribonuclease Cas6 [Streptococcus thermophilus]
MKKLVFTFKRIDHPAQDLAVKFHGFLMEQLDSDYVDYLHQQQTNPYATKVIQGKENTQWVVHLLTDDHEDKVFMTLLQIK
EVSLNDLPKLSVEKVEIQELGADKLLEIFNSEENQTYFSIIFETPTGFKSQGSYVIFPSMRLIFQSLMQKYGRLVENQPE
IEEDTLDYLSEHSTITNYRLETSYFRVHRQRIPAFRGKLTFKVQGAKTLKAYVKMLLTFGEYSGLGMKTSLGMGGIKLEE
RKDpython 实现
import hashlib
filename = "./P06_000_000_001.fasta"
with open(filename) as f:
lines = f.readlines()
res = []
for line in lines:
if not line.startswith(">"):
res.append(line.strip())
# md5 the res string
md5 = hashlib.md5()
md5.update(''.join(res).encode('utf-8'))
print(md5.hexdigest().upper())使用 jsw_bio
import jsw_bio as bio
fasta_str = open(filename).read()
bio.ncbi_fasta2md5(fasta_str)
# CCD30D35DEF426190203046E3E71F214