ncbi: ipg 爬虫相关问题记录
关于数据提取过程中的 ncbi_session_id 过期问题处理
🕐
RestClient 配置
@id = 558016069
@format = json
### get nsbi_sid
POST https://www.ncbi.nlm.nih.gov/sviewer/ipg/ipg.cgi
user-agent: Mozilla/5.0 zgrab/0.x
db=ipg&solr=1&id={{id}}
### 557992326 - NOT
POST https://www.ncbi.nlm.nih.gov/sviewer/ipg/ipg.cgi
content-type: application/x-www-form-urlencoded
cookie: ncbi_sid=CE8BBFD72B412CA1_2139SID; NCBI-PHID=CE8BBFD72B3E9E1100000000085B02D1.m_13;
user-agent: Mozilla/5.0 (Macintosh; Intel Mac OS X 10_15_7) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/102.0.0.0 Safari/537.36
query=omitHeader%3Dfalse%26wt%3D{{ format }}%26indent%3Dtrue%26rows%3D50%26start%3D0%26q%3Drecordset%253A{{ id }}%2520NOT%2520product_i%253A1%26sort%3Dpriority_i%2520asc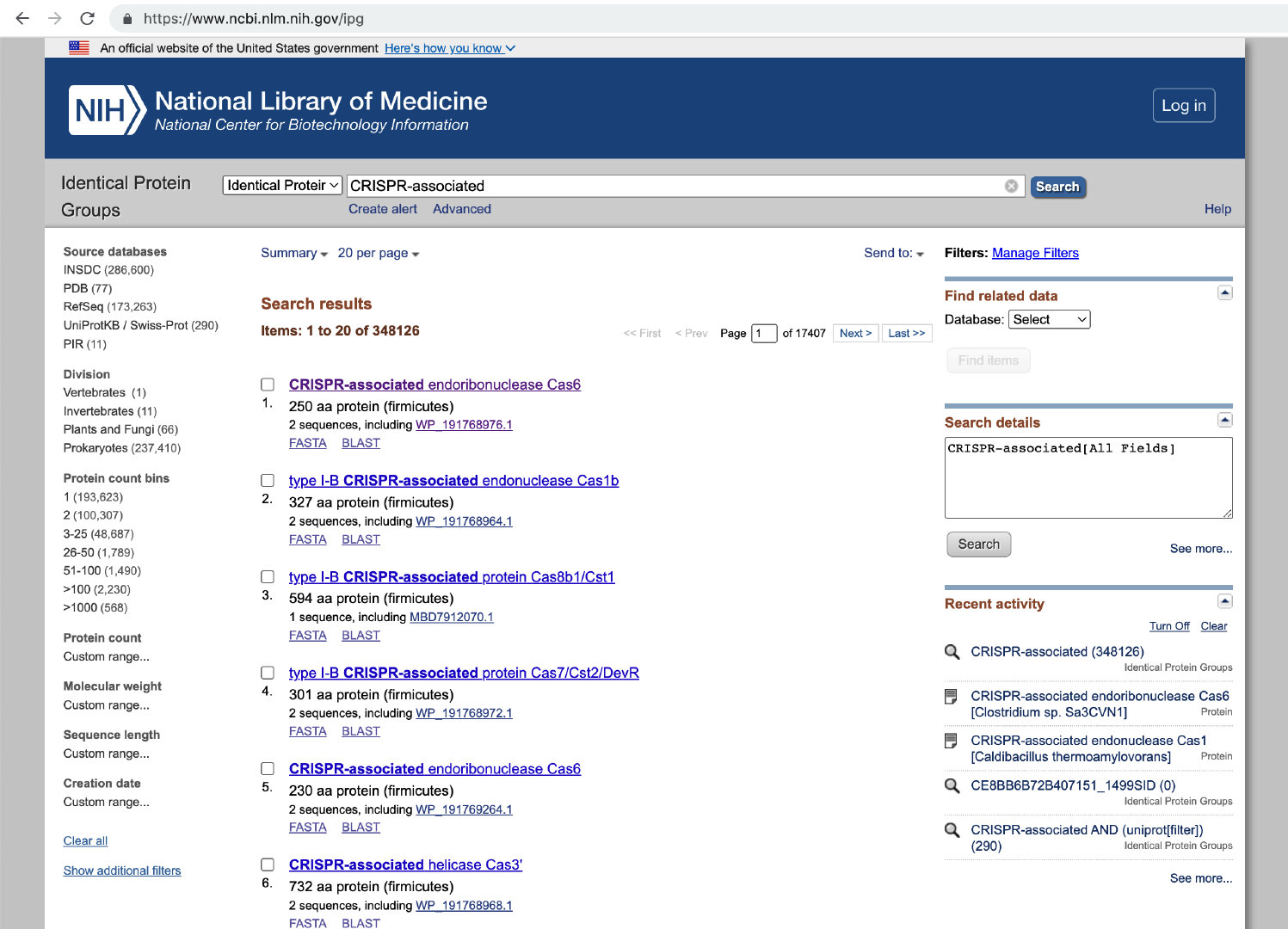
解释
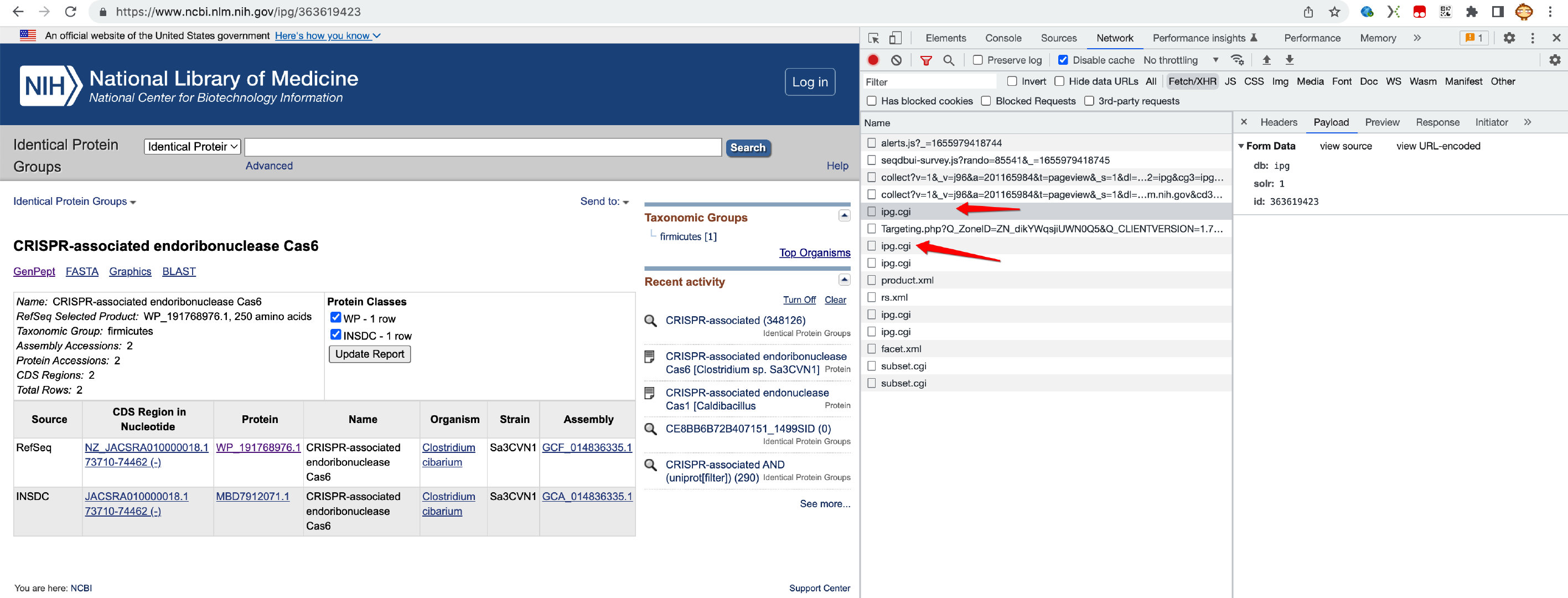
- 请求1会发送id的相关信息过去,产生一个
ncbi_session_id - 后面再用这个id,就可以顺利取到相关数据
采集列表
以 “CRISPR-associated“ 关键词,采集这个关键词列表的数据
### GET qky
GET https://www.ncbi.nlm.nih.gov/ipg?term=CRISPR-associated
### simple
# <input name="EntrezSystem2.PEntrez.DbConnector.LastQueryKey" sid="1" type="hidden" value="1">
POST https://www.ncbi.nlm.nih.gov/ipg
cookie: ncbi_sid=CE8A78F52B64F241_1325SID;
term=CRISPR-associated+&EntrezSystem2.PEntrez.Ipg.Ipg_ResultsPanel.Ipg_DisplayBar.PageSize=20&EntrezSystem2.PEntrez.Ipg.Ipg_ResultsPanel.Entrez_Pager.CurrPage=1&EntrezSystem2.PEntrez.DbConnector.LastQueryKey=1&EntrezSystem2.PEntrez.DbConnector.Cmd=PageChanged原理
- 先发送”
CRISPR-associated“ 的 GET 请求- 生成临时的 session(会有一段时间有效)
- 同时返回2个关相的KEY
ncbi_sid=CE8A78F52B64F241_1325SID- EntrezSystem2.PEntrez.DbConnector.
LastQueryKey
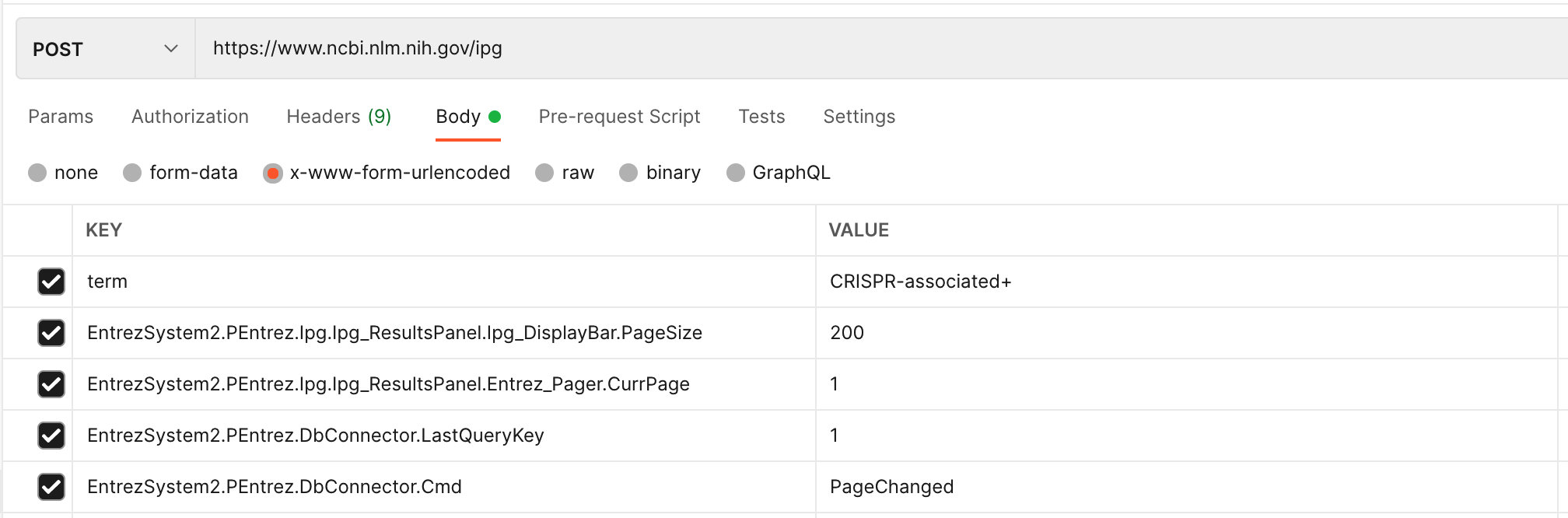
- 根据第1步生成的2个Key,再发送第2步POST请求,分页获取数据
term:CRISPR-associated+
EntrezSystem2.PEntrez.Ipg.Ipg_ResultsPanel.Ipg_DisplayBar.PageSize:200 # 每页取
EntrezSystem2.PEntrez.Ipg.Ipg_ResultsPanel.Entrez_Pager.CurrPage:1 # 当前页
EntrezSystem2.PEntrez.DbConnector.LastQueryKey:1 # 需要动态取, 随 session_id 动态取
EntrezSystem2.PEntrez.DbConnector.Cmd:PageChanged # 除第1页之外的都得带,所以,通用情况下,都带上这一个,否则分页不生效Python 的实现
import requests
import jsw_nx as nx
from bs4 import BeautifulSoup
# ========== step1: reset ncbi_session_id/last_query_key =======
keyword = 'CRISPR-associated'
url = f'https://www.ncbi.nlm.nih.gov/ipg?term={keyword}'
res1 = requests.get(url)
soup1 = BeautifulSoup(res1.text, 'html.parser')
last_query_key_el = soup1.select_one(
'[name="EntrezSystem2.PEntrez.DbConnector.LastQueryKey"]')
# string: 1/2/3/4
last_query_key = last_query_key_el.attrs['value']
# string: CE8C42592B6531E1_1815SID
ncbi_session_id = res1.headers.get('NCBI-SID')
# ======== step2: get pagination data =====
post_data = {
'term': 'CRISPR-associated',
'EntrezSystem2.PEntrez.Ipg.Ipg_ResultsPanel.Ipg_DisplayBar.PageSize': 20,
'EntrezSystem2.PEntrez.Ipg.Ipg_ResultsPanel.Entrez_Pager.CurrPage': 1,
'EntrezSystem2.PEntrez.DbConnector.LastQueryKey': last_query_key,
'EntrezSystem2.PEntrez.DbConnector.Cmd': 'PageChanged'
}
res2 = requests.post("https://www.ncbi.nlm.nih.gov/ipg", data=post_data, headers={
'cookie': f'ncbi_sid={ncbi_session_id}',
})
soup2 = BeautifulSoup(res2.text, 'html.parser')
print(
soup2.select_one('title'),
soup2.select_one('[name="ncbi_pageno"]'),
soup2.select_one('[name="ncbi_pagesize"]')
)利用 jsw-bio
- 安装
- 采集 ids
pip install jsw-bio -U按分页采集ids
import jsw_bio as bio
inst1 = bio.NcbiSearchIpg(term="crispr")
inst2 = bio.NcbiSearchProtein(term="crispr")
ids1 = inst1.get(1, size=20)
ids2 = inst2.get(1, size=20)
print(ids1, ids2)